Authors: Zhilin Huang, Ling Yang, Zaixi Zhang, Xiangxin Zhou, Yu Bao, Xiawu Zheng, Yuwei Yang, Yu Wang, Wenming Yang
Published on: January 15, 2024
Impact Score: 7.6
Arxiv code: Arxiv:2402.18583
Summary
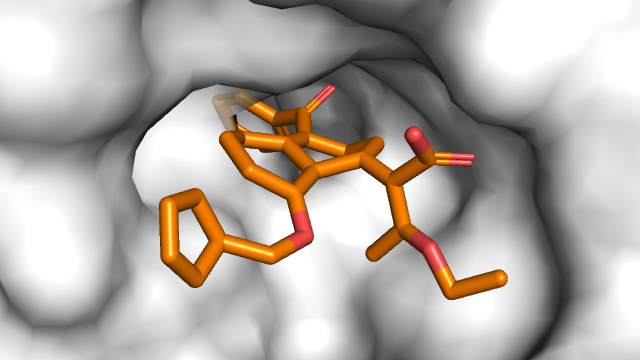
- What is new: Introduces Binding-Adaptive Diffusion Models (BindDM) for capturing protein-ligand interactions in 3D molecule generation.
- Why this is important: Existing 3D deep generative models for structure-based drug design struggle to accurately capture essential protein-ligand interactions.
- What the research proposes: BindDM extracts essential binding sites (subcomplex) and uses SE(3)-equivariant neural networks to infuse binding interaction information into the molecule generation process.
- Results: BindDM generates molecules with more realistic 3D structures and higher binding affinities, achieving up to -5.92 Avg. Vina Score.
Technical Details
Technological frameworks used: BindDM
Models used: SE(3)-equivariant neural networks
Data used: CrossDocked2020 dataset
Potential Impact
Pharmaceutical companies, biotech startups in drug discovery, companies in molecular simulation software.
Want to implement this idea in a business?
We have generated a startup concept here: BindOptics.

Leave a Reply