Authors: Alex Morehead, Jianlin Cheng
Published on: February 08, 2023
Impact Score: 8.77
Arxiv code: Arxiv:2302.04313
Summary
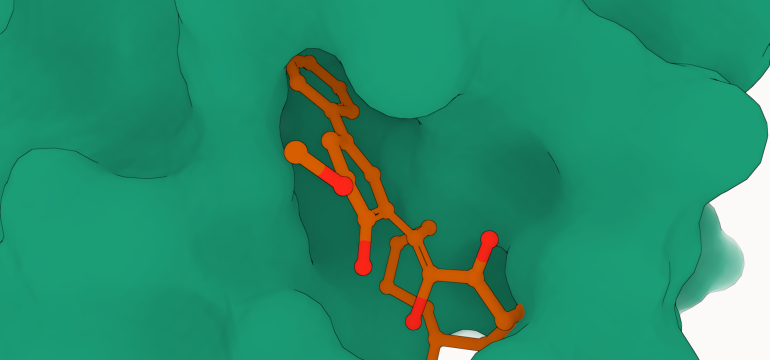
- What is new: The introduction of the Geometry-Complete Diffusion Model (GCDM) for 3D molecule generation, with significant improvements over existing methods.
- Why this is important: Existing methods for generating 3D molecules using denoising diffusion probabilistic models and equivariant graph neural networks fail to learn critical geometric and physical properties, limiting their effectiveness on large molecule datasets.
- What the research proposes: The GCDM leverages a geometry-complete denoising process that enables the generation of realistic and stable large 3D molecules, outperforming previous models on both the QM9 and GEOM-Drugs datasets.
- Results: GCDM not only generates high-quality 3D molecules but also extends to designing molecules for specific protein pockets and optimizing the geometry and chemical composition of molecules for desired properties.
Technical Details
Technological frameworks used: Denoising diffusion probabilistic models (DDPMs), Equivariant Graph Neural Networks (GNNs)
Models used: Geometry-Complete Diffusion Model (GCDM)
Data used: QM9 dataset, GEOM-Drugs dataset
Potential Impact
Pharmaceuticals, computational biology companies, and businesses in the computer vision sector could both be affected and benefit from advancements offered by GCDM.
Want to implement this idea in a business?
We have generated a startup concept here: MolDesign AI.



Leave a Reply